Predicting Antibiotic Resistance Using One-shot Learning
Using Siamese networks to predict antibiotic resistance
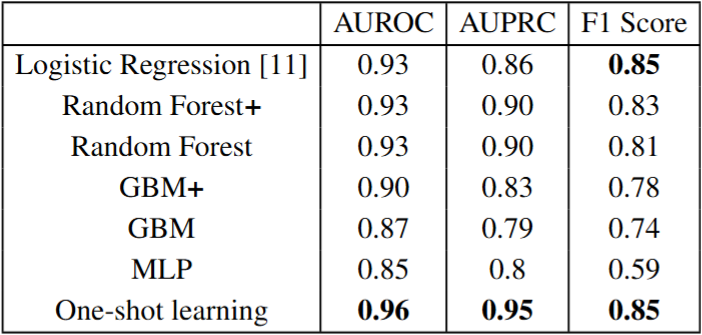
Even as new drugs continue to be developed, the potential of antimicrobial resistance (AMR) threatens the prevention and treatment of various microbe-caused diseases. According to the Center for Disease Control and Prevention, there are more than 2.8 million antibiotic-resistant infections each year resulting in over 35,000 deaths. Because of this, the ability to identify and analyze the biological mechanisms behind AMR is a pressing issue in the healthcare field. Recent advances have made data annotating bacterial genetic regions associated with AMR available through sources such as PATRIC. Furthermore, machine learning methods have previously made use of this data to then predict bacterial resistance based on genotype. In particular, Mahé and Tournoud made use of k-mers and stability selection to predict bacterial resistance against various drugs based on whole bacterial genomes. This project proposes that deep learning models can be used to improve on the results of previous machine learning methods. In particular, the lack of data suggests that this task might benefit from one-shot or few-shot learning. Unlike traditional deep learning classification models, one-shot learning does not emphasize how the sample should be classified. Instead, the model learns a feature embedding which separates the samples of different classes from each other. Given the common difficulty of collecting large corpora of data, various model types have been applied to one and few-shot learning tasks. For this reason, our siamese network-based method is beneficial to the task of predicting antibiotic resistance.
This project was part of the 2952G Deep Learning in Genomics course.