Large Image Segmentation with GNN
Memory Efficient Semantic Segmentation of Large Microscopy Images Using Graph-based Neural Networks
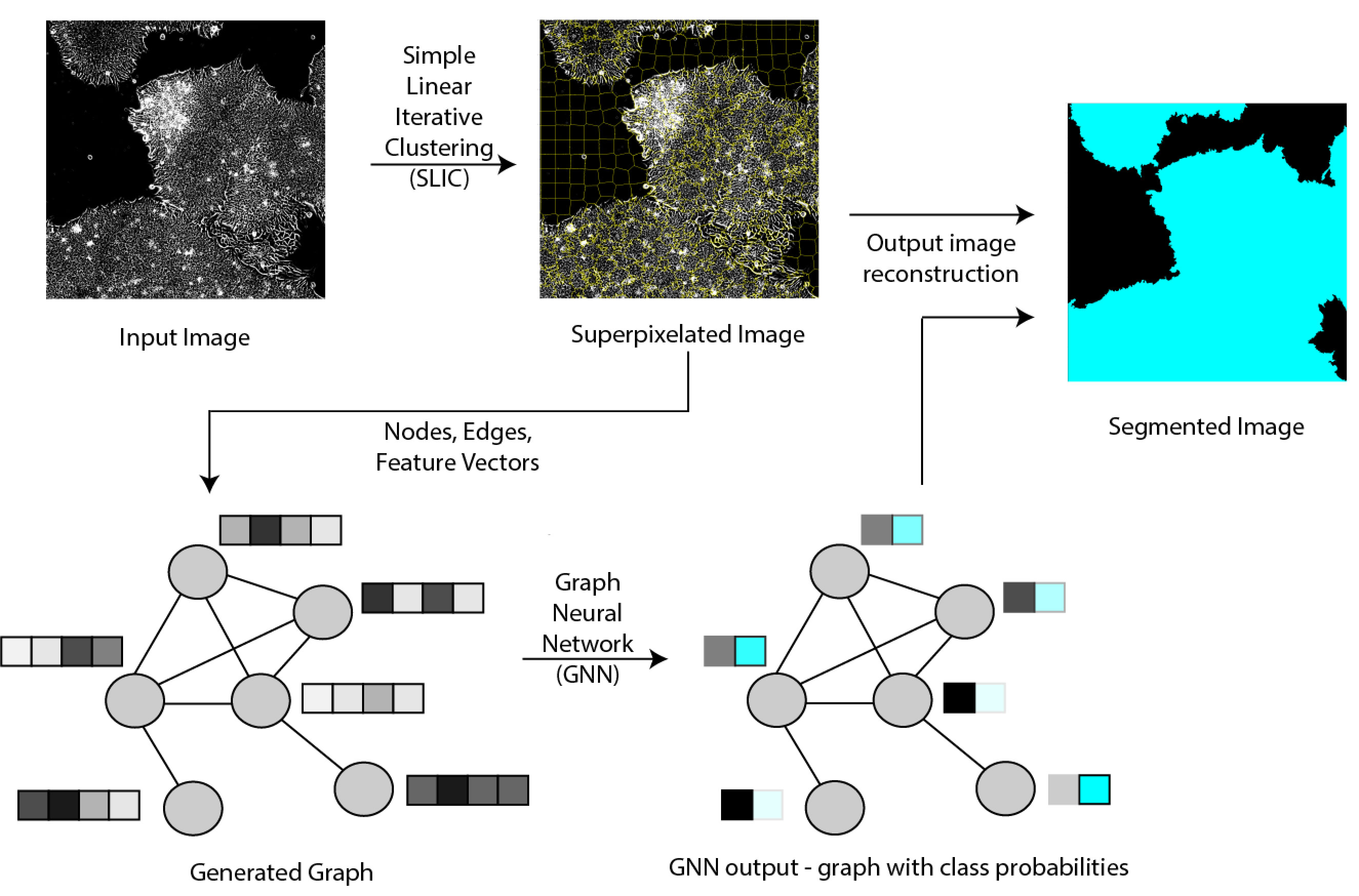
This project is a graph neural network (GNN)-based framework applied to large-scale microscopy image segmentation tasks. While deep learning models, like convolutional neural networks (CNNs), have become common for automating image segmentation tasks, they are limited by the image size that can fit in the memory of computational hardware. In a GNN framework, large-scale images are converted into graphs using superpixels (regions of pixels with similar color/intensity values), allowing us to input information from the entire image into the model. By converting images with hundreds of millions of pixels to graphs with thousands of nodes, we can segment large images using memory-limited computational resources. We compare the performance of GNN- and CNN-based segmentation in terms of accuracy, training time, and required graphics processing unit (GPU) memory. Based on our experiments with microscopy images of biological cells and cell colonies, the GPU-based segmentation used one to three orders-of-magnitude fewer computational resources with only a -2% to +0.3% change in accuracy. Furthermore, errors due to superpixel generation can be reduced by either using better superpixel generation algorithms or increasing the number of superpixels, thereby allowing for improvement in the GNN framework’s accuracy. This tradeoff between accuracy and computational cost over CNN models makes the GNN framework attractive for many large-scale microscopy image segmentation tasks in biology.