STING
Spatial Transcriptomics cluster Inference with Nested Graph neural networks
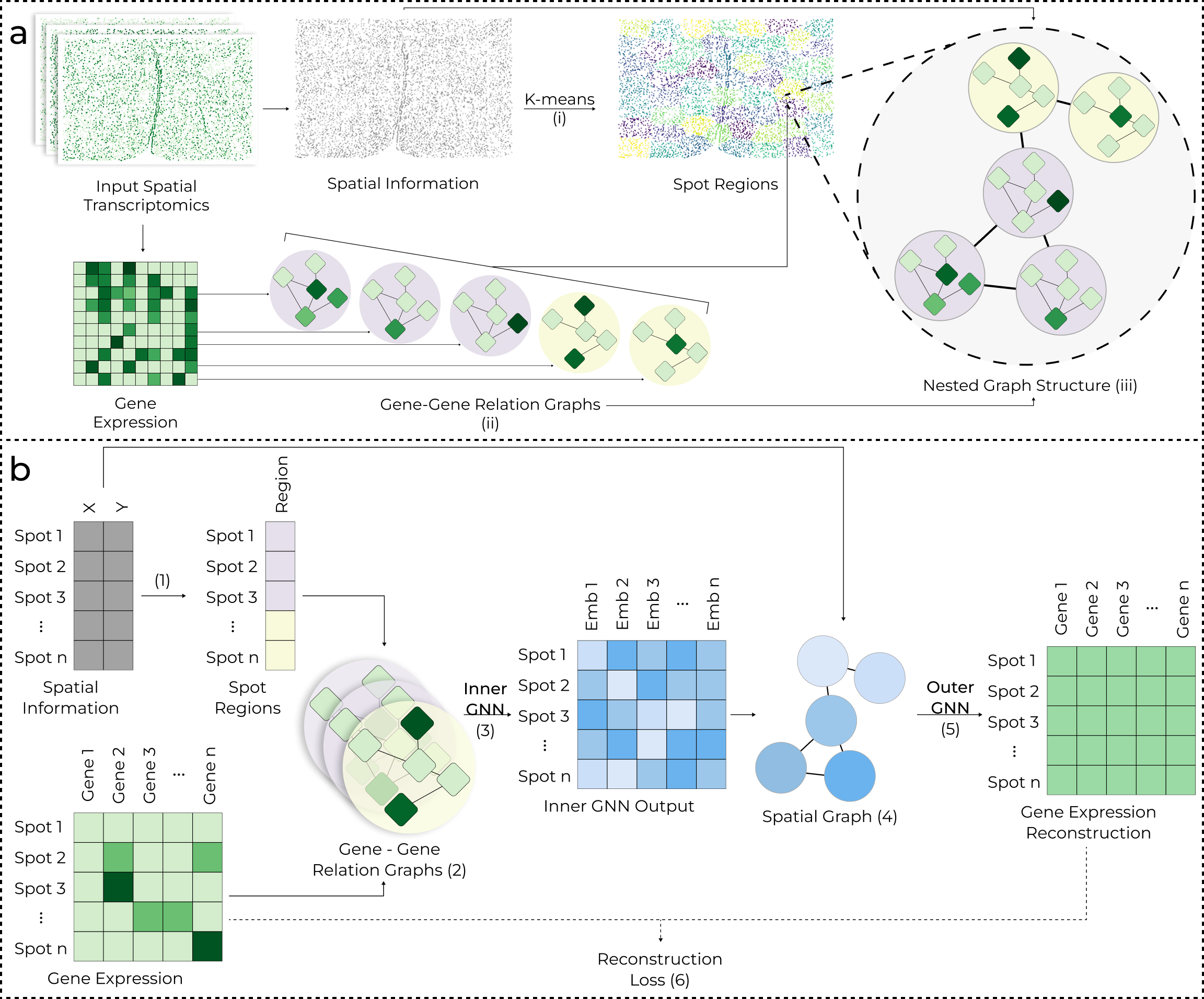
Spatial transcriptomics clustering improves our understanding of tissue environments and disease pathology. While existing clustering methods have proven useful, they overlook spot-level gene-gene relations that can provide meaningful biological context. To address this, we introduce a novel nested graph-based neural network (GNN) approach, STING, for spatial transcriptomic clustering analysis. Unlike existing state-of-the-art techniques that only train on graphs generated from the spatial proximity of spots, STING simultaneously models gene-gene and spatial relations. Experiments on 45 samples across 9 datasets and 5 spatial transcriptomics technologies show that STING outperforms two existing state-of-the-art techniques, MuCoST and GraphST, with an average 12.37% and 17.09% improvement in clustering accuracy measured by Adjusted Rand Index (ARI), respectively. Moreover, qualitative results show that STING has more spatially coherent clusters and is better aligned with manually annotated ground truth clusters. Ablation studies demonstrate the significance of spot-level gene-gene interactions. Further experiments demonstrate STING’s ability to identify key gene-gene interactions in tissues, enabling its use in biological hypothesis generation.
GitHub [Publication TBA]